Research
I am a structural biologist studying HIV infection on the molecular level at Stanford University. In my research, I use my expertise in structural biology, biochemistry, and biophysics to reveal the molecular basis of infectious diseases and to expand our understanding of antimicrobial drugs. Right now, I am focusing on uncovering the regulatory role of RNA structures in reverse transcription of the HIV virus. To achieve my aim, I develop structural biology and biophysical tools to track the viral RNA rearrangements inside the HIV virus during reverse transcription. My research vision is to bring the structure of large RNAs to life by tracking their real-time rearrangements in situ and revealing their function in human health and disease.
Past accomplishments
HIV viral RNA structure and function during reverse transcription
I focus on revealing the regulatory role of RNA structures in reverse transcription of the HIV virus, at my postdoctoral research with Professors Joseph Puglisi and Elisabetta Viani Puglisi at Stanford. The role of RNA structures in early steps of reverse transcription are poorly understood. Specifically, the complex process of reverse transcription initiation and the first event of strand transfer. My analysis of reverse transcription initiation, from the structural biology perspective, highlights the fascinating questions and current challenges to understanding this process (Krupkin et al, Current Opinion in Structural Biology, 2020). My analysis of the mutation pattern at the 5’UTR of HIV viral RNA reveals structure-function relationship (J Gen Virol, 2023). I set up a system to prepare a synthetic full-length viral RNA in the lab, which allows me to start studying the conformational landscape of HIV viral RNA during reverse transcription (in progress).
- Krupkin M*, Jackson LN*, Ha B*, Puglisi EV. “Advances in understanding the initiation of HIV-1 reverse transcription”. Current Opinion in Structural Biology (2020). Link.
- Nouhin J, … Krupkin M, Puglisi JD, Puglisi EV, Shafer RW. Human immunodeficiency virus 1 5’-leader mutations in plasma viruses before and after the development of reverse transcriptase inhibitor-resistance mutations. J Gen Virol (2023). Link.
Synthetic biology
I contributed to development of new synthetic tRNAs for synthetic biology, by building the model of the synthetic tRNA, data analysis, and identification of which nucleotides to mutate to allow for efficient translation (Prabhakar et al. 2022).
- Prabhakar A, … Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic ́A, Ehrenberg M, Puglisi EV, So ̈ll, and Puglisi J. Uncovering translation roadblocks during the development of a synthetic tRNA. Nucleic Acids Res (2022). Link. Journal cover.
Mucus in health and disease
During my time at MIT, I focused on developing a system to study the structure and function of mucus by using purified mucin gels and microfluidics (Wagner, Krupkin et al, Biomacromolecules, 2023). My work set the foundation to study the interaction between mucins and human galactin-3 (JACS Au, 2024), and intelectin-2 (submitted) using mucus gel that I reconstituted from mucins I purified. This work lays the basis for clinical research of the permeability of various mucus layers, which plays a key role in health and disease. Furthermore, my system can be used to study drug delivery.
- Wagner CE*, Krupkin M*, Smith-Dupont KB*, Wu CM, Bustos NA, Witten J, Ribbeck K. Comparison of Physicochemical Properties of Native Mucus and Reconstituted Mucin Gels. Biomacromolecules (2023). Link. *First co-authors.
- Diehl RC, …, Krupkin M, Ribbeck K, Kulik HJ, and Kiessling LL. A CH-π Interaction Is Required for Human Galectin-3 Function. JACS Au (2024). Link.
- Dugan AE, …, Krupkin M, Cárcamo-Oyarce G, Ribbeck K, Xavier RJ, Bevins CL, and Kiessling LL. Intelectin-2 is a broad-spectrum antimicrobial lectin. BioRxiv Link.
Ribosomal antibioticis
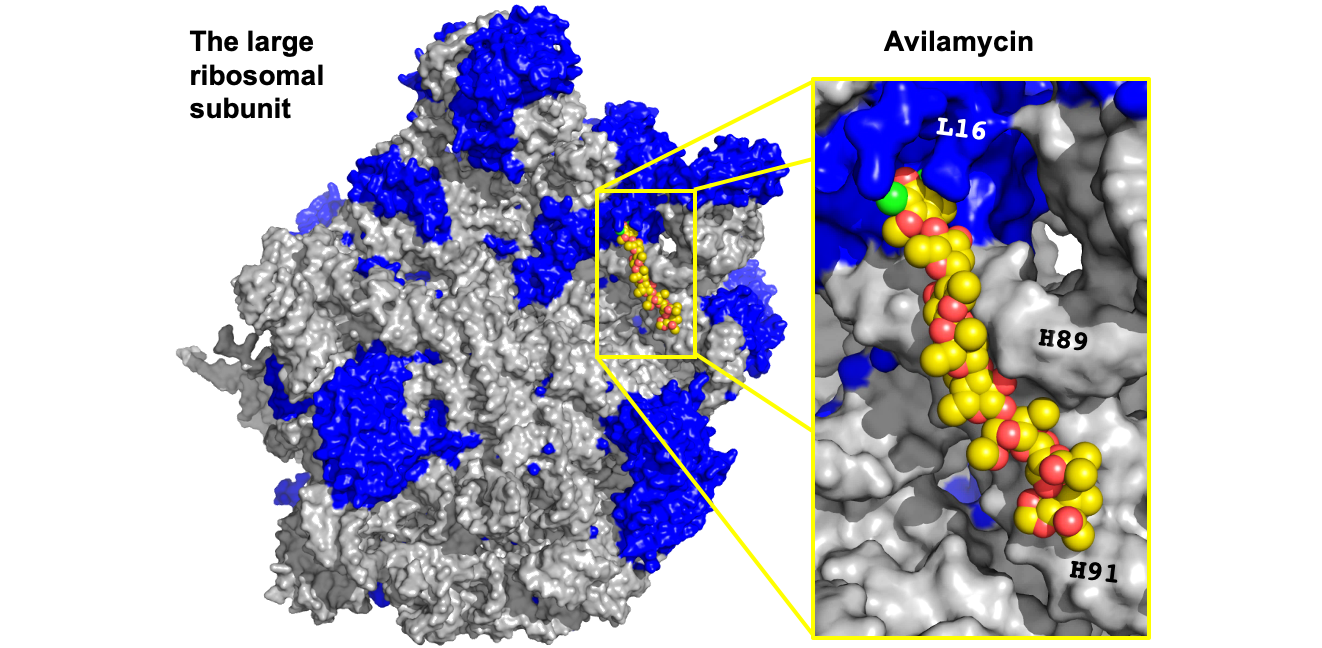
During my PhD research with Professor Ada Yonath at the Weizmann Institute of Science, I expanded our understanding of the molecular mechanism of avilamycin, a large polysaccharide ribosomal antibiotic (Krupkin et al, PNAS 2016). For this purpose, I solved the crystal structure of avilamycin, both free and in complex with the bacterial ribosome. First, I repurposed cutting edge crystallographic machinery and software to determine the structure of the full-length free avilamycin, pushing the abilities to determine small molecule structures. Then, by analyzing the structure of the bound drug, I revealed how avilamycin is utilizing the tRNA binding and the accommodation path to hinder the ribosomal activity. In addition, I uncovered how ribosomal mutations can change the binding pocket and create bacterial resistance to the drug, and elucidated how the differences between the human and the bacterial ribosome constitute the basis for avilamycin selectivity, an essential feature for effective antibiotics.
In addition to studying the orthosomycin ribosomal antibiotics, I contributed to studying macrolides and pleuromutilins antibiotics (Matzov et al 2017, Wekselman et al 2017) and uncovering ribosomal structures from the life-threatening pathogen Staphylococcus aureus(Eyal et al 2015, Eyal et al 2016), by crystal handling, sample freezing, data collection, and data analysis. These studies expand our understanding of species-specific resistance and drug inhibition mechanisms of multi drug resistance pathogens.
- Krupkin M*, Wekselman I*, Matzov D, Eyal Z, Diskin Posner Y, Rozenberg H, Zimmerman E, Bashan A, Yonath A. “Avilamycin and evernimicin induce structural changes in rProteins uL16 and CTC that enhance the inhibition of A-site tRNA binding”. Proc Natl Acad Sci USA (2016). Link.
- Eyal Z*, Matzov D*, Krupkin M, Wekselman I, Paukner S, Zimmerman E, Rozenberg H, Bashan A, Yonath A. “Structural insights into species-specific features of the ribosome from the pathogen staphylococcus aureus”. Proc Natl Acad Sci USA (2015). Link.
- Eyal Z*, Matzov D*, Krupkin M, Paukner S, Riedl R, Rozenberg H, Zimmerman E, Bashan A, and Yonath A. “A novel pleuromutilin antibacterial compound, its binding mode and selectivity mechanism”. Scientific Reports (2016). Link.
- Auerbach-Nevo T, … Krupkin M, … Yonath A. “Ribosomal antibiotics: Contemporary challenges”. Antibiotics (2016). Link.
- Matzov D, … Krupkin M, Zimmerman E, Rozenberg H, Bashan A, Fridman M, Yonath A. “Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus”. Nucleic Acids Res (2017). Link.
- Wekselman I, … Krupkin M, Rozenberg H, Bashan A, Friedlander G, Kjeldgaard J, Ingmer H, Lindahl L, Zengel JM, Yonath A. “The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel”. Structure (2017). Link.
Ribosomal origins
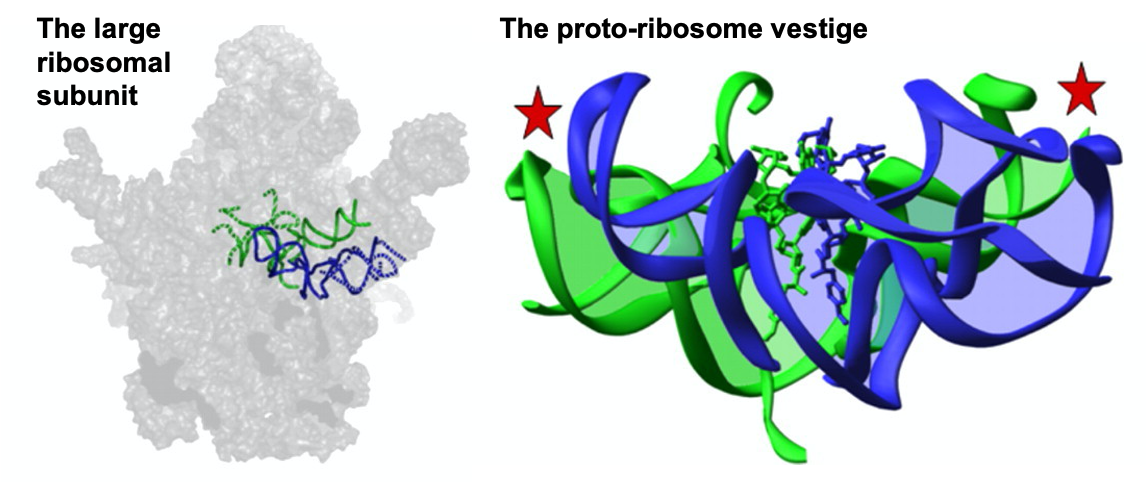
I studied the ability of the ribosomal RNA fragments to form peptide bonds, during my graduate studies with Professor Ada Yonath (Krupkin et al, Philos Trans R Soc Lond B Biol Sci 2011, Krupkin et al 2014). I proposed aa novel hypothesis that the ribosome has evolved from the pre-proto-ribosome, a molecular machine capable of performing essential tasks in the RNA world. This pre-proto- ribosome was a molecular machine capable of performing essential tasks in the RNA world, which was later snatched for producing proteins by the amino acids invaders. To explore this hypothesis, I reconstructed the ribosomal active site from short RNA strands I designed. I showed that these RNA molecules were able to dimerize (Bose et al, NAR 2022), as I hypothesized from the ribosome structure and was supported by computational approaches (Huang, Krupkin, et al. PNAS 2013). Furthermore, these molecules were able to perform peptide-bond formation! My research contributes to the understanding of the emergence and evolution of protein translation; a process central to all known life forms.
- Krupkin M, Matzov D, Tang H, Metz M, Kalaora R, Belousoff MJ, Zimmerman E, Bashan A, Yonath A. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos Trans R Soc Lond B Biol Sci (2011). link. Research highlight: Link.
- Krupkin M, Bashan A, Yonath A. (2014) “Glimpse into the Origin of Life: What was First, the Genetic Code or its Products, the Proteins?” in “Why does Evolution Matter? The Importance of Understanding Evolution”, G. Trueba, ed. (Cambridge Scholars Publishing), p. 87-100. Link.
- Bose T, Fridkin G, Davidovich C, Krupkin M, Dinger N, Falkovich AH, Peleg Y, Agmon I, Bashan A, Yonath A. “Origin of life: proto ribosome forms peptide bonds and links RNA and protein dominated worlds”. Nucleic Acids Res (2022). Link. Media: Nature, PhysOrg.
- Huang L, Krupkin M, Bashan A, Yonath A, Massa L. “Protoribosome by quantum kernel energy method”. Proc Natl Acad Sci USA (2013). Link.
- Davidovich C, … Krupkin M, Zimmerman E, Bashan A, Yonath A. “The proto-ribosome : An ancient nano-machine for peptide bond formation”. Isr J Chem. (2010). Link. Journal cover.
Other Interests
I am devoted to promoting science education and outreach. In my spare time, I love to do yoga. 
HIV and Structural Biology news
Read more about what’s new in HIV research here and here. Read more about what’s new in structural biology here.
